The Missing Link in Pre-Clinical Genome Editing Workflows
Currently, the “gold standard” genome editing workflows only produce one data type: in-cell editing events. With this single parameter, there is just one outcome to optimize around. Discovery programs typically focus on guide RNAs (gRNA) redesign or transfection conditions to maximize sensitivity and selectivity to troubleshoot CRISPR-Cas genome editing. However, valuable information about CRISPR-Cas performance is hidden in the biochemical steps upstream of a genome editing event. Suppose a genome editing experiment doesn’t work or generates off-target editing. It could be that gRNA binding to Cas enzyme is low affinity or binding to off-target DNA is more favorable than on-target binding. Yet, gathering this data requires biochemical interrogation rather than iterative in-cell editing experiments. Collecting data on the steps that precede DNA cleavage can give genome editing programs additional parameters to optimize, providing more insight into CRISPR-Cas activity and, ultimately, improving in-cell editing outcomes.
Below, we peek inside the black box of CRISPR-Cas structure and function, looking at each step in the editing process, from gRNA binding to DNA cleavage, and discuss opportunities for optimization.
- Structure of Cas Proteins and gRNA
- A Step-by-Step Walkthrough of CRISPR-Cas Activity
- Opportunities for CRISPR-Cas Optimization
- References
Structure of Cas Proteins and gRNA
Each Cas enzyme has its eccentricities (i.e., PAM sequence specificity, structure, etc.), and cleavage activity may differ. Cas9 is the most studied Cas enzyme, so its molecular underpinnings are the most understood and will be the focus of our discussion.
Cas9 Protein Structure
The Cas9 protein can be broken down into 6 distinct protein domains, each with its role in the DNA cleavage process:1-3
- Rec I and II domains: These are responsible for binding the gRNA, with the Rec I domain doing the majority of the heavy lifting.
- Bridge-Helix domain: Helps initiate cleave following binding to complementary DNA sequence.
- PAM Interacting domain (PID): The protospacer adjacent motif (PAM) sequence is downstream of the target DNA. The PID is what determines the sequence specificity of the PAM and mediates the initial recognition of potential target DNA. The sequence differs across different Cas enzymes; in the case of Cas9, it’s an NGG.
- HNH and RuvC domains: These are nuclease domains, each of which can cut a single strand of DNA. Together, they facilitate double-stranded DNA cleavage. Cas9 can be turned into a nicking enzyme through mutagenesis of either of these domains, only cutting one DNA strand. Both domains can also be mutated to turn Cas9 into dead Cas9 (dCas9), which is used for sequence-specific DNA binding or gene-specific expression or repression.
gRNA Structure
A gRNA is a single RNA strand with a 5′ complementarity region (for targeting a specific DNA sequence), a downstream tetraloop, and two to three small stem-loops.
A Step-by-Step Walkthrough of CRISPR-Cas Activity
When using CRISPR-Cas systems, we typically think about DNA cleavage, either nicking or double-stranded break (DSB). But DNA cleavage occurs in a 5-step process that involves i) gRNA-Cas9 ribonucleoprotein (RNP) complex formation, ii) DNA scanning and PAM recognition, iii) base pairing, iv) complete binding, and v) DNA cleavage.
Here’s a closer look at what happens at each step. As mentioned above, different Cas systems have subtle differences in their mechanisms of action, but many landmark structure-function papers published use Cas9, which is what we’ll review below.
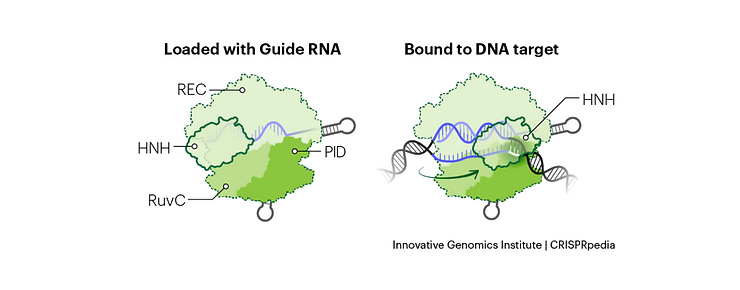
Step #1: gRNA-Cas9 RNP Complex Formation
The Cas9 protein is inactive when not bound to a gRNA with the appropriate structures mentioned above.2 When Cas9 binds to a gRNA, the tetraloop of the gRNA is bound by the Rec I and II domains, inducing a conformational change in the Cas9 enzyme and the formation of an active RNP complex.1 The PID and RuvC domain also binds to the 3′ stem-loop structures.1
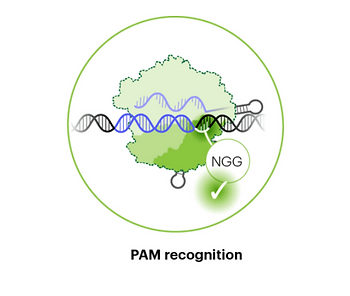
Step #2: PAM Recognition
Once active, the Cas9 RNP searches for its cognate PAM sequence. Recognition of the PAM is mediated by the PID domain and occurs through random collisions with DNA.4 Once a potential target sequence is recognized, there is also local sliding with a 20 nucleotide region to test other possible DNA sequences in nearby loci.5 Recently, an energy-based model was developed to account for this local sliding and could more accurately predict on- and off-target editing than other in silico models.6
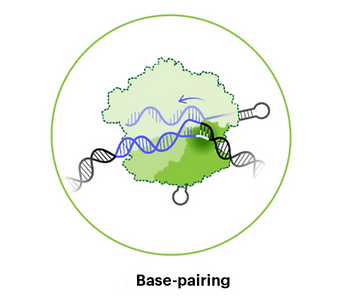
Step #3: Unwinding Complementary DNA and Base Pairing
Following PAM recognition, the adjacent DNA is unwound and binds to complementary RNA within the gRNA (called the “seed region”).4 After this initial binding, the rest of the complementary region within the guide binds to the target DNA sequence. During recognition of the target DNA strand, the non-target strand is held separately by the Cas9 enzyme. If binding is not entirely complementary, the Cas9 RNP may unbind and move on to interrogate other sequences.
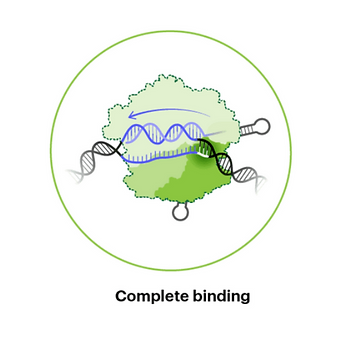
Step #4: Completing Binding
If a PAM is spaced correctly next to a DNA sequence complementary to the gRNA, then the Cas9 protein is primed to cleave both DNA strands. Some mismatches in specific regions of the gRNA-DNA heteroduplex are tolerated, leading to off-target binding and downstream cleavage.7
Step #5: Target DNA Cleavage
Once the entire gRNA (or most of the gRNA) is bound to the target DNA sequence, there is a conformational change, and the HNH domain cleaves the DNA strand bound to the gRNA, 3 nucleotides upstream of the PAM sequence. Next, the RuvC domain cleaves the non-target DNA strand.
Opportunities for CRISPR-Cas Optimization at CRISPR QC™
In a cell, the generation of a nicked strand or a DSB signals DNA repair machinery to take over, which is how insertion-deletion (indel) mutations or homologous recombination with exogenous DNA can be achieved. This activity is preceded by the biochemical steps outlined above, which can require fine-tuning for efficient, on-target editing in a cell.
At CRISPR QC™, we provide deep analytics about the biochemical steps underlying CRISPR-Cas function. Knowing which CRISPR-Cas activities can be improved gives you the insight and opportunity for performance optimization and, therefore, a higher probability of driving a successful CRISPR program.
Talk to our technical experts today to see how you can leverage our platform to generate valuable insights into your CRISPR-Cas activity.
References:
- Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935-949. doi:10.1016/j.cell.2014.02.001.
- Jinek M, Jiang F, Taylor DW, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. doi:10.1126/science.1247997.
- Kanafi MM, Tavallaei M. Overview of advances in CRISPR/deadCas9 technology and its applications in human diseases. Gene. 2022;830:146518. doi:10.1016/j.gene.2022.146518.
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62-67. doi:10.1038/nature13011.
- Globyte V, Lee SH, Bae T, Kim JS, Joo C. CRISPR/Cas9 searches for a protospacer adjacent motif by lateral diffusion. EMBO J. 2019;38(4):e99466. doi:10.15252/embj.201899466.
- Corsi GI, Qu K, Alkan F, Pan X, Luo Y, Gorodkin J. CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context. Nat Commun. 2022;13(1):3006. Published 2022 May 30. doi:10.1038/s41467-022-30515-0.
Fu BX, St Onge RP, Fire AZ, Smith JD. Distinct patterns of Cas9 mismatch tolerance in vitro and in vivo. Nucleic Acids Res. 2016;44(11):5365-5377. doi:10.1093/nar/gkw417