Customer noticed the gRNA selections as predicted by in-silico algorithms didn’t lead to optimal editing outcomes. Sequencing data only told them that the edits didn’t occur, but not why.
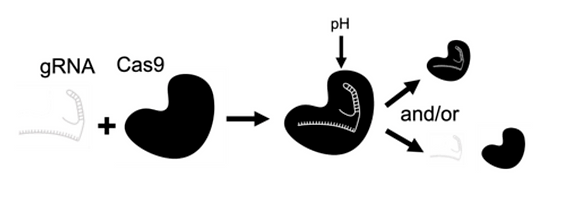
pH was determined to be a causal factor in their editing, something in-silico algorithms couldn’t emulate.
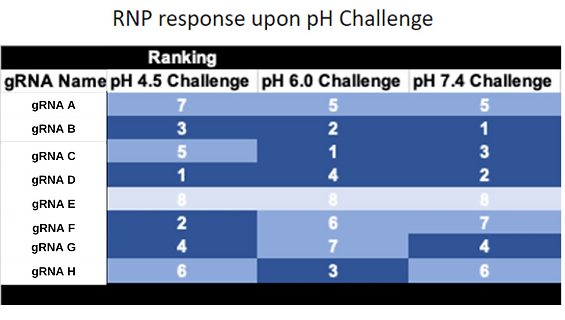
Customer was able to use our in-vitro data to select effective gRNAs. This greatly reduced the amount of cell work needed to produce a repeatable edit.
(858) 285 -4122
info@crisprqc.com
8949 Kenamar Drive, Suite 101
San Diego, CA 92121, USA
© 2025 CRISPR QC™, Inc.