Increase gene editing and biomanufacturing success through quantitative measurement of gRNA design efficiency, RNP formation, and complex stability.
The root cause of your CRISPR editing failures may begin with inefficient Cas-gRNA ribonucleoprotein (RNP) complex assembly—but you may have a hard time proving it. Traditional methods, which rely on indirect measurements or post-experiment analysis, fail to provide real-time insights into RNP formation. This lack of clarity frequently leads to months of frustrating guesswork and troubleshooting, stalling research progress and draining valuable time and resources.
Do you have additional workflow challenges you’d like to overcome? Learn more below.
Illuminate RNP assembly dynamics using quantitative real-time measurement of relative binding kinetics and stability via our CRISPR Analytics Platform™. This precision approach allows you to pinpoint bottlenecks in complex formation, enabling data-driven modifications that streamline your CRISPR workflow.
Explore our case study showcasing how customers leveraged the CRISPR Analytics Platform™ to overcome challenges, from identifying optimal gRNA designs when in silico predictions fell short, to resolving inconsistencies in multiplex gene editing by selecting gRNAs with similar displacement rates for uniform editing frequencies.
Leverage in vitro CRISPR analytics to optimize:

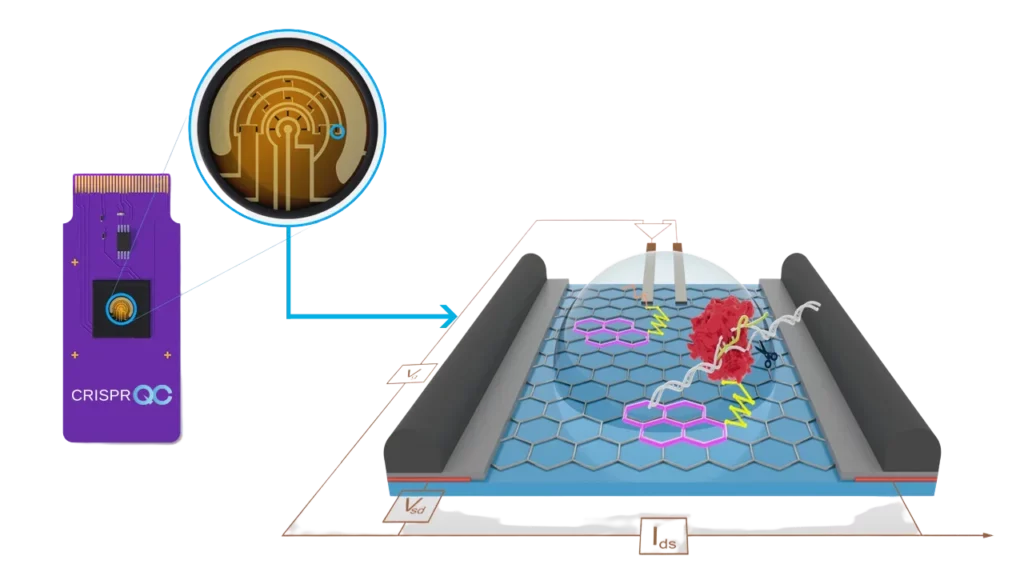
Our cutting-edge CRISPR-Chip™ Biosensor combines the sensitivity of a graphene-based field-effect transistor with the specificity of CRISPR by immobilizing Cas proteins onto the graphene layer.
The sensitivity of graphene’s conductivity to biomolecular recognition on its surface endows the platform with the unique capacity to quantitatively assess binding interactions or cleavage reactions in real time. Together, these elements deliver actionable insights, empowering your CRISPR-Cas biochemistry investigations with unparalleled accuracy and efficiency.
© 2025 CRISPR QC™, Inc.